The knowledge of complex three-dimensional folds of RNA structures is essential to understand the
increasing number of their biological functions. Both, X-ray crystallography and NMR spectroscopy,
are principal experimental methods to deliver RNA structural models at high resolution. However,
there is a great demand in the biological research community to envisage new, yet experimentally
inaccessible three-dimensional RNA structures to address particular questions on the structure-function
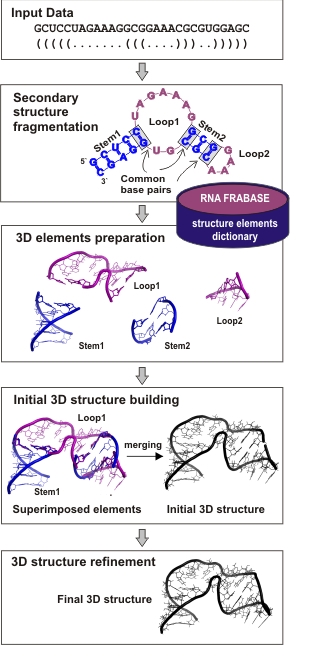
relationship. Here, we present RNAComposer (version 1.0), an advanced tool for modeling of RNA structures.
It's aim is to build atomic-resolution 3D models of large RNAs, based on their secondary structures,
using the RNA FRABASE dictionary relating
RNA secondary structure and tertiary structure elements.
The general idea of RNAComposer is based on the following processing path:
Top ↑

RNAComposer works in two modes:
Top ↑
- interactive mode
Interactive mode is mainly intended for those, who want to test the capabilities of the system. It allows to run RNAComposer for a single sequence (up to 500 nt long) and its secondary structure topology to build one output 3D model. A user is provided the information about processing status in the real time. Pdb file with the generated model is available for download immediately after the end of modeling. This file can be sent via email, to the user request. In addition, JSmol visualization of the model is provided on the webpage. More about this mode... - batch mode
In the batch mode, available for registered users, RNAComposer can compose of up to 1000 RNA structure 3D-models based on a set of 100 different secondary structures, provided by single user at a time. Although, in principle, the method itself allows to build models for RNA sequences of unlimited length, in the batch mode we have set the server threshold to strands of up to 500 nt residues. The data entered by the user as a single batch is queued in the system and served according to the queueing algorithm. Pdb file(s) and log file(s) generated for the batch are available for download immediately after the end of modeling and can be accessed from the user workspace. According to the account settings these files can be sent via email to the given address. More about this mode...
Interactive mode is designed for users who want to predict a single structure for
a short sequence (up to 500 nts). Users are recommended to provide their own secondary
structure as input data. However, in this mode, it is possible to use one of the
applications for RNA secondary structure prediction which have been incorporated
into the RNAComposer pipeline.
A secondary structure introduced into RNAComposer define a single task in the system. Processing a task in interactive mode results in generating a single RNA 3D structure.
a. Input data
Input data in interactive mode define one task and consist of: task identifier, sequence, and - optionally - secondary structure topology or application for secondary structure prediction. Each information is provided in a separate line. Thus, a typical input should be formatted in the following way:
Optionally, in any place of the entry, the user can insert a comment line starting from "#".
First line indicates where task definition begins. It must be placed before the specification of a sequence and a secondary structure topology. This line is followed by the exact definition of the structure, in which user can specify:
If the 3rd line of the entry remains empty and the method is not selected from the list, CentroidFold is applied by default.
Dot-bracket notation
RNAComposer uses dot-bracket notation to represent RNA secondary structure topology. Usually, in this notation:
User should provide an input secondary structure topology encoded in dot-bracket. However, we support also CT and BPSEQ notations by providing converters between these formats and dot-bracket in the Tools page.
Applications for RNA secondary structure prediction
For user convenience, the following applications for RNA secondary structure prediction have been incorporated into RNAComposer pipeline:
Accepted characters
The following characters are accepted when entered into the task entry box:
Top ↑
A secondary structure introduced into RNAComposer define a single task in the system. Processing a task in interactive mode results in generating a single RNA 3D structure.
a. Input data
Input data in interactive mode define one task and consist of: task identifier, sequence, and - optionally - secondary structure topology or application for secondary structure prediction. Each information is provided in a separate line. Thus, a typical input should be formatted in the following way:
| line | type | contents |
| 1st line | obligatory | ">" followed by a unique task identifier |
| 2nd line | obligatory | RNA sequence in the one-letter format |
| 3rd line | optional | secondary structure topology in dot-bracket notation / application for RNA secondary structure prediction |
Optionally, in any place of the entry, the user can insert a comment line starting from "#".
First line indicates where task definition begins. It must be placed before the specification of a sequence and a secondary structure topology. This line is followed by the exact definition of the structure, in which user can specify:
- only the sequence.
- the sequence and the secondary structure topology.
- the sequence and the application for RNA secondary structure prediction.
If the 3rd line of the entry remains empty and the method is not selected from the list, CentroidFold is applied by default.
Dot-bracket notation
RNAComposer uses dot-bracket notation to represent RNA secondary structure topology. Usually, in this notation:
- an unpaired nucleotide is represented as a dot "."
- a base pair is represented as a pair of opening (left) and closing (right) brackets, i.e. "(" and ")".
User should provide an input secondary structure topology encoded in dot-bracket. However, we support also CT and BPSEQ notations by providing converters between these formats and dot-bracket in the Tools page.
Applications for RNA secondary structure prediction
For user convenience, the following applications for RNA secondary structure prediction have been incorporated into RNAComposer pipeline:
- CentroidFold (default selection)
- ContextFold
- CONTRAfold
- IPknot
- RNAfold
- RNAstructure.
Accepted characters
The following characters are accepted when entered into the task entry box:
| type of contents | accepted characters / strings | additional information |
| strand identifier | >, A-Z, a-z, 0-9 |
line must start with > max length: 10 characters |
| sequence | A, C, G, U, T | |
| secondary structure topology | . ( ) [ ] { } < > | |
| secondary structure prediction | CentroidFold, ContextFold, CONTRAfold, IPknot, RNAfold, RNAstructure | case insensitive |
| comment line | any characters | line must start with # |
b. Output data
In the interactive mode the user obtains the following output:
After running RNAComposer for a task, a user is taken to the task progress information page, where the succeeding steps of structure modeling are announced. The information about the result of each step and its computing time is released in the real time, due to a progress of computation. Just after the end of modeling, a pdf file with the resulting structure is available for download and visualized with JSmol. The pdf can be sent via email to the address provided by the user with entry data.
Top ↑
In the interactive mode the user obtains the following output:
- information about task processing phases,
- pdf file with a generated structure,
- JSmol visualization of a model.
- a log with 2D structure fragmentation
After running RNAComposer for a task, a user is taken to the task progress information page, where the succeeding steps of structure modeling are announced. The information about the result of each step and its computing time is released in the real time, due to a progress of computation. Just after the end of modeling, a pdf file with the resulting structure is available for download and visualized with JSmol. The pdf can be sent via email to the address provided by the user with entry data.
c. Example use
Three examples, ready to be loaded, are available to the user. Example 1 shows a query which should be provided to model a structure of HIV-2 DIS RNA hairpin. Example 2 concerns encoding RNA structure with a pseudoknot, which is also supported by RNAComposer. Example 3 presents the way to run RNAComposer when one does not have a secondary structure and decides to use one of the tools incorporated into the system.
The following tables show graphical and text (in the RNAComposer accepted format) representation of each example input.
Top ↑
Three examples, ready to be loaded, are available to the user. Example 1 shows a query which should be provided to model a structure of HIV-2 DIS RNA hairpin. Example 2 concerns encoding RNA structure with a pseudoknot, which is also supported by RNAComposer. Example 3 presents the way to run RNAComposer when one does not have a secondary structure and decides to use one of the tools incorporated into the system.
The following tables show graphical and text (in the RNAComposer accepted format) representation of each example input.
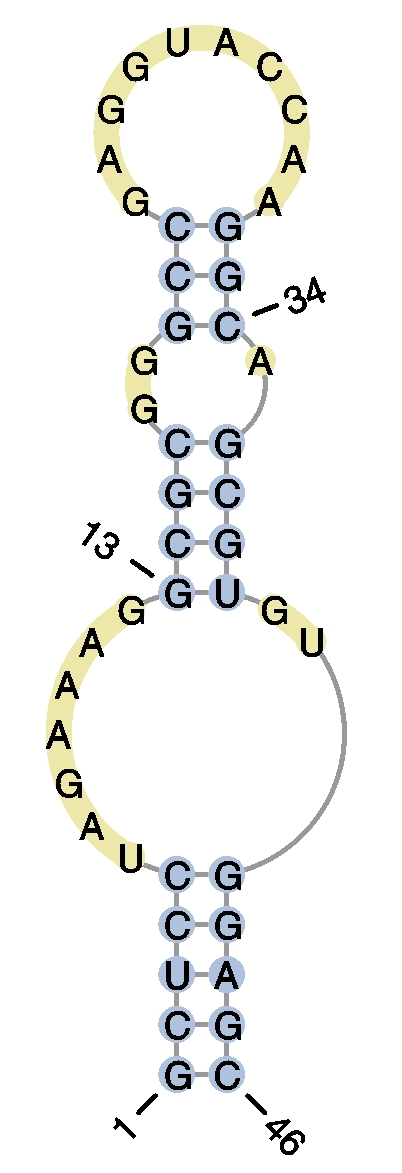
- Example 1
user query query graphical representation #HIV-2 DIS RNA hairpin
>example1
GCUCCUAGAAAGGCGCGGGCCGAGGUACCAAGGCAGCGUGUGGAGC
(((((.......((((..(((..........))).))))..)))))
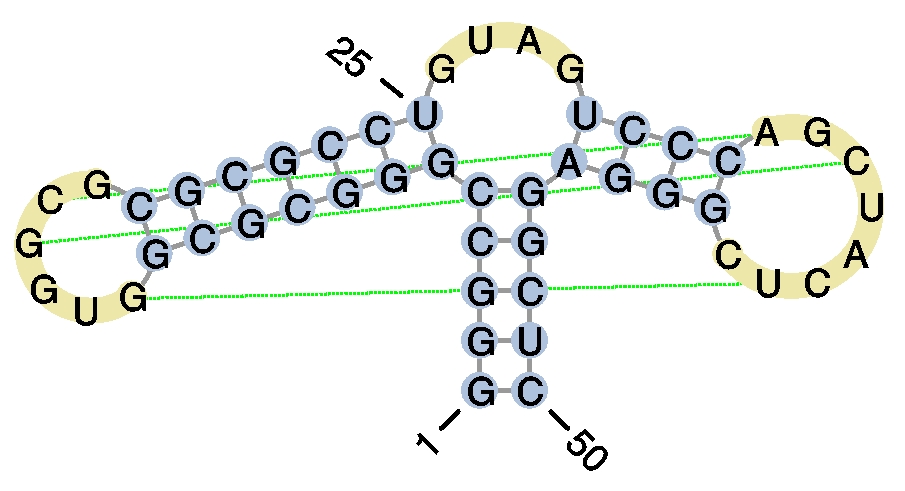
- Example 2
user query query graphical representation #pseudoknot, Alu domain of the mammalian signal recognition particle (PDB: 1E8O)
>example2
GGGCCGGGCGCGGUGGCGCGCGCCUGUAGUCCCAGCUACUCGGGAGGCUC
((((((((((((..[[[.)))))))....((((.]]..]..)))))))))
- Example 3
user query #RNA sequence for a construct containing hammerhead ribozyme and GNRA loop motifs
#RNAfold - to generate secondary structure
>example3
ACCAGCUGAUGAGUCCCAAAUAGGACGAAACGCCGUAAGGCGUCCUGGU
RNAfold
Batch mode is available only for registered users.
To enter this mode login to the system is required.
Batch mode has been designed for those who perform large-scale modeling. Input sequence length in this mode is limited up to 500 nts. 1-10 secondary structures can be provided for a single sequence (at least one is required). User is allowed to provide of up to 10 sequences at a time. Such a multiple input is called a batch. One batch is composed of tasks (a task is associated with one secondary structure). Processing a task in the batch mode results in generating either one or multiple output (i.e. 1-10 RNA 3D structures depending on the user requirements).
a. Input data
The following input data are accepted in the batch mode:
Batch
Batch can be either entered directly to the entry box or uploaded from a file. In both cases the same data format must be followed. A batch should consist of 1-10 sections. Each section concerns one sequence and contains: an identifier, a sequence, and - optionally - 1-10 definitions of secondary structure topology associated with the same sequence. Each information should be provided in a separate line. A single batch section should be formatted in the following way:
First line indicates where the section begins. It must be placed before the specification of a sequence. The secondary structure topology - if provided - should be encoded in the dot-bracket notation. For those having their secondary structures in CT or BPSEQ, appropriate converters to dot-bracket are available in the Tools page. If neither secondary structure topology nor the name of application for secondary structure prediction is provided, the structure is predicted by CentroidFold.
Optionally, in any place of the batch, user can insert a comment line starting with "#".
Characters accepted in the batch
The following characters are accepted in the batch entry box and batch file:
Batch example
Three examples, ready to be loaded, are available to the user. Example 1 shows a query which defines a structure of tetrahymena ribozyme (1X8W, chain B) with 247 nts log sequence. One secondary structure is provided in this example. Example 2 presents the way to format a batch including several different secondary structure topologies for one sequence. A 120 nts long sequence of 5S rRNA E.Coli taken from 2AWB (chain A, mutant 12C>12A) serves as the example. Example 3 concerns runing RNAComposer for a multi-sequence batch.
The following tables show each example input.
Filtering 3D structure elements repository
During preparation of 3D structure elements that compose the 3D model, respective elements are selected from the associated structural repository. User can define constraints (filters) to be applied during 3D elements search. Four filtering options are available after clicking the Filter 3D structure elements repository checkbox:
3D structure minimization restraints
User can restrain the process of 3D structure minimization by providing restraints for atom distances or torsion angles. This option is enabled if the user clicks Define 3D structure minimization restraints checkbox under batch entry box and selects one of checkboxes that appear thereafter:
Atom distance restraints
In order to enter atom distance restraints one should fist click Define 3D structure minimization restraints, and next check Add atom distance restraints checkbox. This enables entering the data into a separate entry box.
Atom distance restraints can be either entered directly to the restraints entry box or uploaded from a file. In both cases the same data format must be followed. User provides a set of distance restraints concerning one or more secondary structures. More than one set of restraints can be provided at a time. Subsequent sections containing set descriptions should be separated by an empty line. A single section (set of restraints) should be formatted as follows:
Atom distance restraints example
Torsion angle restraints
In order to enter torsion angle restraints one should fist click Define 3D structure minimization restraints, and next check Torsion angle restraints checkbox. This enables entering the data into a separate entry box.
Torsion angle restraints can be either entered directly to the restraints entry box or uploaded from a file. In both cases the same data format must be followed. User provides a set of angle restraints concerning one or more secondary structures. More than one set of restraints can be provided at a time. Subsequent sections containing set descriptions should be separated by an empty line. A single section (set of restraints) should be formatted as follows:
Torsion angle restraints example
Batch mode has been designed for those who perform large-scale modeling. Input sequence length in this mode is limited up to 500 nts. 1-10 secondary structures can be provided for a single sequence (at least one is required). User is allowed to provide of up to 10 sequences at a time. Such a multiple input is called a batch. One batch is composed of tasks (a task is associated with one secondary structure). Processing a task in the batch mode results in generating either one or multiple output (i.e. 1-10 RNA 3D structures depending on the user requirements).
a. Input data
The following input data are accepted in the batch mode:
- batch with sequences (obligatory) and secondary structures (optional),
- constraints for 3D structure elements repository search (optional),
- restraints concerning 3D structure minimization (optional),
- 3D structure elements to be inserted into the predicted RNA model (optional).
Batch
Batch can be either entered directly to the entry box or uploaded from a file. In both cases the same data format must be followed. A batch should consist of 1-10 sections. Each section concerns one sequence and contains: an identifier, a sequence, and - optionally - 1-10 definitions of secondary structure topology associated with the same sequence. Each information should be provided in a separate line. A single batch section should be formatted in the following way:
| line | type | contents |
| 1st line | obligatory | ">" followed by a unique identifier |
| 2nd line | obligatory | RNA sequence in the one-letter format |
| 3rd-12th line | optional |
secondary structure topologies in the dot-bracket notation or name of the application for secondary structure prediction (one secondary structure or application name per line) |
First line indicates where the section begins. It must be placed before the specification of a sequence. The secondary structure topology - if provided - should be encoded in the dot-bracket notation. For those having their secondary structures in CT or BPSEQ, appropriate converters to dot-bracket are available in the Tools page. If neither secondary structure topology nor the name of application for secondary structure prediction is provided, the structure is predicted by CentroidFold.
Optionally, in any place of the batch, user can insert a comment line starting with "#".
Characters accepted in the batch
The following characters are accepted in the batch entry box and batch file:
| type of contents | accepted characters / strings | additional information |
| strand identifier | >, A-Z, a-z, 0-9 |
line must start with > max length: 10 characters |
| sequence | A, C, G, U, T | |
| secondary structure topology | . ( ) [ ] { } < > | |
| secondary structure prediction | CentroidFold, ContextFold, CONTRAfold, IPknot, RNAfold, RNAstructure | case insensitive |
| comment line | any characters | line must start with # |
Batch example
Three examples, ready to be loaded, are available to the user. Example 1 shows a query which defines a structure of tetrahymena ribozyme (1X8W, chain B) with 247 nts log sequence. One secondary structure is provided in this example. Example 2 presents the way to format a batch including several different secondary structure topologies for one sequence. A 120 nts long sequence of 5S rRNA E.Coli taken from 2AWB (chain A, mutant 12C>12A) serves as the example. Example 3 concerns runing RNAComposer for a multi-sequence batch.
The following tables show each example input.
- Example 1
user query #Tetrahymena ribozyme (PDB: 1X8W; chain B)
>example1
GACCGUCAAAUUGCGGGAAAGGGGUCAACAGCCGUUCAGUACCAAGUCUCAGGGGAAACUUUGAGAUGGCCUUGCAAAGGGUAUGGUAAUAAGCUGACGGACAUGGUCCUAACCGCGCAGCCAAGUCCUAAGUCAACAGGAGACUGUUGAUAUGGAUGCAGUACACAGACUAGAUGUCGGCCGGGGAAGAUGUAUUCUUCUCAUAAGGUAUAGUCGGACCUCUCCCGAAAGGGAGUUGGAGUACUCG
(.(.(.(....((((((...(.((((.....(((.((((.(((..(((((((((....)))))))))..((.......))....)))......)))))))....)))).)..)).))))((...((.(...((((((((....))))))))..).))...))...[.[[[[[...).).).).((((((.......))))))........]]]]]](.((.(((((....)))))..))......).
- Example 2
user query #5S rRNA E.Coli (PDB: 2AWB; chain A; mutant 12C>12A)
>example2
UGCCUGGCGGCAGUAGCGCGGUGGUCCCACCUGACCCCAUGCCGAACUCAGAAGUGAAACGCCGUAGCGCCGAUGGUAGUGUGGGGUCUCCCCAUGCGAGAGUAGGGAACUGCCAGGCAU
((((((((((.....((((((((.....((((((...(.....)...))))..))....)))))).)).((.......((((((((...)))))))).......))...)))))))))).
((((((((((.........((((....)))).((((((((((.....(((....)))...(((((.......))))).))))))))))((((..(((....))))))).)))))))))).
((((((((((.....((((((((.((....((((.............))))....))..)))))).)).((.((....((((((((...))))))))....)).))...)))))))))).
- Example 3
user query #pre-miRNA
>hsa_mir_132
ACCGUGGCUUUCGAUUGUUACUGUGGGAACUGGAGGUAACAGUCUACAGCCAUGGUCG
(((((((((...((((((((((............))))))))))...)))))))))..
(((((((((...((((((((((((....))....))))))))))...)))))))))..
(((((((((...((((((((((.(((...)))..))))))))))...)))))))))..
>hsa_mir_136
ACUCCAUUUGUUUUGAUGAUGGAUUCUUAUGCUCCAUCAUCGUCUCAAAUGAGUCU
((((.(((((...((((((((((.........))))))))))...)))))))))..
(((.((((((...((((((((((.........))))))))))...)))))))))..
>hsa_mir_139
UCUACAGUGCACGUGUCUCCAGUGUGGCUCGGAGGCUGGAGACGCGGCCCUGUUGGAGU
(((((((.((.((((((((((((...........)))))))))))))).))).))))..
(((((((.((.((((((((((((...........)))))))))))))).)))).)))..
((.((((.((.((((((((((((...........)))))))))))))).)))).))...
>hsa_mir_591
AGACCAUGGGUUCUCAUUGUAAUAGUGUAGAAUGUUGGUUAACUGUGGACUCCCUGGCUCUGU
((((((.((((((.((((((((((........)))))..))).)).)))).)).))).)))..
((((((.((((((.(((((...(((..(...)..)))..))).)).)))).)).))).)))..
((((((.((((((.((...((((..............))))..)).)))).)).))).)))..
Filtering 3D structure elements repository
During preparation of 3D structure elements that compose the 3D model, respective elements are selected from the associated structural repository. User can define constraints (filters) to be applied during 3D elements search. Four filtering options are available after clicking the Filter 3D structure elements repository checkbox:
- Exclude selected PDB structures
After clicking this checkbox, user can provide PDB identifiers (case-insensitive)
of all structures that should be dismissed during 3D structure elements search.
Every PDB id should be given in a separate line. Other accepted separators of identifiers
are ',' (comma) and ';' (semicolon). The list of identifiers
can be uploaded from file or entered directly to the entry box (the box is
displayed after clicking Exclude selected PDB structures).
- Use X-ray determined structures only
If this option is selected, only these 3D structure elements are accepted by the search procedure which derive from X-ray determined structures. Additionally, in the Set resolution threshold entry box, user defines the maximum value of resolution threshold (default = 3.0A) that should be satisfied by source X-ray structures (and thus, by the 3D structure elements themselves).
- Generate A-RNA-based single strands
If this option is selected, all single stranded fragments of the predicted 3D structure are generated by NAB procedure (Case et al., 2015), based on the input sequence and the template structure of A-RNA.
- Generate A-RNA-based double helices
If this option is selected, all double helices within the predicted 3D structure are generated by NAB procedure (Case et al., 2015), based on the input sequence and the template structure of A-RNA.
3D structure minimization restraints
User can restrain the process of 3D structure minimization by providing restraints for atom distances or torsion angles. This option is enabled if the user clicks Define 3D structure minimization restraints checkbox under batch entry box and selects one of checkboxes that appear thereafter:
Atom distance restraints
In order to enter atom distance restraints one should fist click Define 3D structure minimization restraints, and next check Add atom distance restraints checkbox. This enables entering the data into a separate entry box.
Atom distance restraints can be either entered directly to the restraints entry box or uploaded from a file. In both cases the same data format must be followed. User provides a set of distance restraints concerning one or more secondary structures. More than one set of restraints can be provided at a time. Subsequent sections containing set descriptions should be separated by an empty line. A single section (set of restraints) should be formatted as follows:
| line | type | contents |
| 1st line | obligatory | section headline containing number(s) of sequence and secondary structure topology |
| 2nd line | obligatory | distance restraints defined for a pair of atoms |
|
3rd line . . . |
optional |
distance restraints defined for another pairs of atoms (one per line) |
- headline
Sequence(s) and secondary structure topologie(s) for which atom distance restraints should be applied are specified in the headline. Headline should have the following format:
<sequence numbers>;<secondary structure topology numbers>
where:
<sequence numbers> is a list of serial numbers of sequences entered in the batch entry box, separated by commas. At least one sequence number is required. A maximum number of specified sequences is 10 (the same as in the batch entry box). Sequences are indexed starting with 1. <secondary structure topology numbers> is a list of serial numbers of secondary structure topologies defined for specified sequence(s), separated by commas. At least one structure number is required. A maximum number of topologies is 10 (same as in the batch entry box, where at most 10 structures are accepted for one sequence). Secondary structure topologies are indexed starting with 1 (separately for each sequence).
Examples of section headline:
1;2
distance restraints defined in the succeeding lines apply to the model(s) built by RNAComposer for the 2nd secondary structure topology provided for the 1st sequence in the batch.
1;1,3
distance restraints defined in the succeeding lines apply to the models built by RNAComposer for the 1st and 3rd secondary structure topologies provided for the 1st sequence in the batch.
1,2;1
distance restraints defined in the succeeding lines apply to the models built by RNAComposer for the 1st secondary structure topology provided for the 1st sequence, and for the 1st secondary structure topology provided for the 2nd sequence in the batch.
2,3;1,3,4
distance restraints defined in the succeeding lines apply to the models built by RNAComposer for the 1st, 3rd and 4th secondary structure topology provided for the 1st sequence, the 1st, 3rd and 4th secondary structure topology defined for the 3rd sequence, and for the 1st, 3rd and 4th secondary structure topology specified under the 4th sequence in the batch.
- distance restraints line
Atom distance restraints are specified under the headline, one line per restraint. Distance restraints line should have the following format:
<residue 1> <atom 1> <residue 2> <atom 2> <distance> <minus> <plus>
i.e. it contains 7 fields, separated by single spaces and having the following meaning:
field meaning format <residue 1>
<residue 2>serial number of the 1st residue
serial number of the 2nd residuemax 4 characters
max 4 characters<atom 1>
<atom 2>name of the atom from the 1st residue
name of the atom from the 2nd residuemax 4 characters
max 4 characters<distance> distance value [Å] %5.2f <minus>
<plus>allowable minus deviation from the distance value [Å]
allowable plus deviation from the distance value [Å]%5.2f
%5.2f
Atom distance restraints example
- Example 1
atom distance restraints description 1;1
4 N1 22 N3 2.85 0.15 0.15
4 N6 22 O4 2.85 0.15 0.15
Specified restraints will be applied to the model built for the 1st secondary structure topology defined for the 1st sequence in the batch.
- Example 2
atom distance restraints description 1;1,2
4 N1 22 N3 2.85 0.15 0.15
4 N6 22 O4 2.85 0.15 0.15
3,4;1
5 N1 32 N3 2.85 0.15 0.15
8 N6 20 O4 2.85 0.15 0.15
First set of restraints will be applied to the models built for the 1st and the 2nd secondary structure topologies defined for the 1st sequence in the batch.
Second set of restraints will be applied to the models built for the 1st secondary structure topologies defined for the 3rd and 4th sequence in the batch.
Torsion angle restraints
In order to enter torsion angle restraints one should fist click Define 3D structure minimization restraints, and next check Torsion angle restraints checkbox. This enables entering the data into a separate entry box.
Torsion angle restraints can be either entered directly to the restraints entry box or uploaded from a file. In both cases the same data format must be followed. User provides a set of angle restraints concerning one or more secondary structures. More than one set of restraints can be provided at a time. Subsequent sections containing set descriptions should be separated by an empty line. A single section (set of restraints) should be formatted as follows:
| line | type | contents |
| 1st line | obligatory | section headline containing number(s) of sequence and secondary structure topology |
| 2nd line | obligatory | restraints defined for 4 atoms involved in particular torsion angle |
|
3rd line . . . |
optional |
restraints defined for 4 atoms involved in another torsion angle (one line contains restrains concerning one torsion angle) |
- headline
Sequence(s) and secondary structure topologie(s) for which torsion angle restraints should be applied are specified in the headline. Headline should have the following format:
<sequence numbers>;<secondary structure topology numbers>
where:
<sequence numbers> is a list of serial numbers of sequences entered in the batch entry box, separated by commas. At least one sequence number is required. A maximum number of specified sequences is 10 (the same as in the batch entry box). Sequences are indexed starting with 1. <secondary structure topology numbers> is a list of serial numbers of secondary structure topologies defined for specified sequence(s), separated by commas. At least one structure number is required. A maximum number of topologies is 10 (same as in the batch entry box, where at most 10 structures are accepted for one sequence). Secondary structure topologies are indexed starting with 1 (separately for each sequence).
Examples of section headline:
1;2
torsion angle restraints defined in the succeeding lines apply to the model(s) built by RNAComposer for the 2nd secondary structure topology provided for the 1st sequence in the batch.
1;1,3
torsion angle restraints defined in the succeeding lines apply to the models built by RNAComposer for the 1st and 3rd secondary structure topologies provided for the 1st sequence in the batch.
1,2;1
torsion angle restraints defined in the succeeding lines apply to the models built by RNAComposer for the 1st secondary structure topology provided for the 1st sequence, and for the 1st secondary structure topology provided for the 2nd sequence in the batch.
2,3;1,3,4
torsion angle restraints defined in the succeeding lines apply to the models built by RNAComposer for the 1st, 3rd and 4th secondary structure topology provided for the 1st sequence, the 1st, 3rd and 4th secondary structure topology defined for the 3rd sequence, and for the 1st, 3rd and 4th secondary structure topology specified under the 4th sequence in the batch.
- torsion angle restraints line
Torsion angle restraints are specified under the headline, one line per restraint. Torsion angle restraints line should have the following format:
<residue 1> <atom 1> <residue 2> <atom 2> <residue 3> <atom 3> <residue 4> <atom 4> <energy> <angle> <range> <centroid> <exp>
i.e. it contains 13 fields, separated by single spaces and having the following meaning:
field meaning format <residue 1>
<residue 2>
<residue 3>
<residue 4>serial number of the 1st residue
serial number of the 2nd residue
serial number of the 3nd residue
serial number of the 4nd residue
max 4 characters
max 4 characters
max 4 characters
max 4 characters
<atom 1>
<atom 2>
<atom 3>
<atom 4>
name of the atom from the 1st residue
name of the atom from the 2nd residue
name of the atom from the 3nd residue
name of the atom from the 4nd residue
max 4 characters
max 4 characters
max 4 characters
max 4 characters
<energy> energy constant value [kcal*mole-1*rad-2] %5.2f <angle> angle value [°] %5.2f <range> angle range %5.2f <centroid> angle centroid value %5.2f <exp> exponent value (optional) %5.2f
Torsion angle restraints example
- Example 1
torsion angle restraints description 1;1
8 P 8 05' 8 C5' 8 C4' 10 -173.5 2.0 2
8 05' 8 C5' 8 C4' 8 C3' 10 50.9 2.0 2
8 C5' 8 C4' 8 C3' 8 03' 10 79.1 2.0 2
Specified restraints will be applied to the model built for the 1st secondary structure topology defined for the 1st sequence in the batch.
- log file with the information about task processing phases and results of each phase,
- pdf file with a generated structure.
- a number of user's batches uploaded to the system,
- detailed information about each batch including: batch identifier, number of sequences in the batch, day & time of an upload, launch and completion by RNAComposer engine,
- result files for completed batches ready for download.
Inserting own 3D structure elements
In the batch mode user has the possibility to interfere in selection of the 3D structure elements that compose the final model. In particular, this option allows the user to replace 3D structure elements selected by the automated routine of RNAComposer. When RNAComposer finishes computation for user input data, it provides the final 3D model and a log file with the results of intermediate computational steps. The log file enumerates i.a. (i) secondary structure elements resulting from input data fragmentation (identifier, sequence and dot-bracket-encoded secondary structure topology are given for every element), and (ii) 3D structure elements associated with their secondary structure counterparts. The user can decide to modify selected parts of the obtained 3D model by providing own 3D structure elements and indicating which elements of originally predicted 3D structure should be substituted.
To do this, one should click Insert own 3D structure elements button and, next, introduce own 3D structure element(s) in PDB format, preceded by the identifier(s) of associated secondary structure element(s). More than one structure element can be substituted at a time. A definition of substitution section should be formatted as follows:
| line | type | contents |
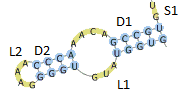
|
| 1st line | obligatory | section headline containing number(s) of sequence and secondary structure topology | |
| 2nd line | obligatory | identifier of the 3D structure element to be substituted (e.g.L1, L2, S1, D2) | |
|
3rd line ... nth line |
obligatory | 3D structure element definition in PDB format | |
| (n+1)th line | optional | identifier of the other 3D structure element to be substituted | |
|
(n+2)th line . . . |
optional | 3D structure element definition in PDB format |
Headline of this section is formatted in the same way as headlines for Atom distance restraints and Torsion angle restraints sections.
Example input is provided below:
1;1 L1 ATOM 214 P U B 25 23.137 11.079 6.937 1.00 19.65 P ATOM 215 OP1 U B 25 23.497 9.696 7.337 1.00 20.82 O ATOM 216 OP2 U B 25 23.891 12.224 7.520 1.00 22.01 O ATOM 217 O5' U B 25 23.218 11.242 5.353 1.00 20.87 O ATOM 218 C5' U B 25 23.077 10.126 4.480 1.00 20.85 C ATOM 219 C4' U B 25 22.510 10.570 3.146 1.00 21.13 C ATOM 220 O4' U B 25 21.116 10.946 3.322 1.00 20.30 O ATOM 221 C3' U B 25 23.131 11.796 2.487 1.00 21.40 C ATOM 222 O3' U B 25 24.296 11.464 1.736 1.00 22.33 O ATOM 223 C2' U B 25 22.014 12.225 1.549 1.00 21.33 C ... ATOM 655 N9 A B 45 17.479 20.602 -0.436 1.00 26.60 N ATOM 656 C8 A B 45 17.059 21.470 0.540 1.00 25.01 C ATOM 657 N7 A B 45 17.170 20.991 1.756 1.00 24.93 N ATOM 658 C5 A B 45 17.696 19.721 1.566 1.00 23.91 C ATOM 659 C6 A B 45 18.060 18.701 2.465 1.00 23.56 C ATOM 660 N6 A B 45 17.959 18.809 3.792 1.00 22.77 N ATOM 661 N1 A B 45 18.544 17.553 1.946 1.00 23.25 N ATOM 662 C2 A B 45 18.662 17.448 0.618 1.00 24.45 C ATOM 663 N3 A B 45 18.367 18.338 -0.327 1.00 24.60 N ATOM 664 C4 A B 45 17.883 19.466 0.221 1.00 24.69 C L2 ATOM 214 P U B 25 23.137 11.079 6.937 1.00 19.65 P ATOM 215 OP1 U B 25 23.497 9.696 7.337 1.00 20.82 O ATOM 216 OP2 U B 25 23.891 12.224 7.520 1.00 22.01 O ... ATOM 664 C4 A B 45 17.883 19.466 0.221 1.00 24.69 C |
Top ↑
b. Output data
In the batch mode user obtains the following output for each model:
After running RNAComposer for a batch, a user can see the progress within the User workspace page. When the batch is finished the results are available for download in a zip file. The file can be sent via email to the address provided by the user in the Account settings.
Top ↑
In the batch mode user obtains the following output for each model:
After running RNAComposer for a batch, a user can see the progress within the User workspace page. When the batch is finished the results are available for download in a zip file. The file can be sent via email to the address provided by the user in the Account settings.
c. User registration
Registration is required for those users who want to run RNAComposer in the batch mode. Interactive mode is available without registration.
To register one should create an account (click Create an account link on the gray area under the main menu). Registration requires providing an email, username (user ID) and password. Note that username must be unique within the system. Check availability button is placed on the registration webpage to help users define a unique ID.
After the account data are submitted to the system (Create an account button clicked), a verification email is sent to the user. Registration is completed when the user clicks the link given in the email. The account is available immediately after verification.
Top ↑
Registration is required for those users who want to run RNAComposer in the batch mode. Interactive mode is available without registration.
To register one should create an account (click Create an account link on the gray area under the main menu). Registration requires providing an email, username (user ID) and password. Note that username must be unique within the system. Check availability button is placed on the registration webpage to help users define a unique ID.
After the account data are submitted to the system (Create an account button clicked), a verification email is sent to the user. Registration is completed when the user clicks the link given in the email. The account is available immediately after verification.
d. Account settings
Once logged into RNAComposer system, user can change account settings including email address and password (to do so select Account settings link from user menu in gray area). The decision on ones datasets (i.e. files including the results of batch processing by RNAComposer) can be also set here. By default, user is notified via email when the results are available (Send email notification without attachements is checked in the Account settings page). The result files are stored in the system for 14 days, available for download from User workspace. The files may be attached to the notification email on user request (check Send email notification with result files attached to activate this option).
Top ↑
Once logged into RNAComposer system, user can change account settings including email address and password (to do so select Account settings link from user menu in gray area). The decision on ones datasets (i.e. files including the results of batch processing by RNAComposer) can be also set here. By default, user is notified via email when the results are available (Send email notification without attachements is checked in the Account settings page). The result files are stored in the system for 14 days, available for download from User workspace. The files may be attached to the notification email on user request (check Send email notification with result files attached to activate this option).
e. User workspace
Logged in user can access his workspace by selecting My workspace from user menu in gray area. The following information is available from the workspace:
To access the information about secondary structures defined in a single batch its identifier should be clicked.
User can delete uploaded batches by selecting a batch (checkbox on the leftmost column should be checked for a batch) and clicking Delete selected button. If not deleted by the user, batches are removed from the system 14 days after completion.
Top ↑
Logged in user can access his workspace by selecting My workspace from user menu in gray area. The following information is available from the workspace:
To access the information about secondary structures defined in a single batch its identifier should be clicked.
User can delete uploaded batches by selecting a batch (checkbox on the leftmost column should be checked for a batch) and clicking Delete selected button. If not deleted by the user, batches are removed from the system 14 days after completion.
Tools page provides support to the users having their RNA secondary
structure encoded in CT (Connect) or BPSEQ format. Since RNAComposer
accepts only dot-bracket notation, we offer an interface to
CT→dot-bracket and BPSEQ→dot-bracket converters. They are
user-friendly shortcuts to functions available in RNApdbee.
Secondary structure in BPSEQ or CT format can be uploaded directly from a file. After conversion, user can save dot-bracket secondary structure into a file ('Save') or send it to the task entry box ('Send to homepage'), where it can be immediately used to run new task.
Example encoding of secondary structure:
Top ↑
Secondary structure in BPSEQ or CT format can be uploaded directly from a file. After conversion, user can save dot-bracket secondary structure into a file ('Save') or send it to the task entry box ('Send to homepage'), where it can be immediately used to run new task.
Example encoding of secondary structure:
| CT format | BPSEQ format | dot-bracket notation | graphical view (by PseudoViewer) |
|
8
1 G 0 2 8 1 2 G 1 3 7 2 3 C 2 4 0 3 4 A 3 5 0 4 5 U 4 6 0 5 6 U 5 7 0 6 7 C 6 8 2 7 |
1 G 8 2 G 7 3 C 0 4 A 0 5 U 0 6 U 0 7 C 2 8 C 1 |
GGCAUUCC
( ( . . . . ) ) |
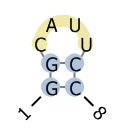
|
RNAComposer is designed to work with most of the available web browsers. The latest
versions of browsers are strongly recommended.
Recommended web browsers
Recommended web browsers
| Operating system | Recommended browser |
| Windows | Microsoft Internet Explorer (11.0 and later), Mozilla Firefox (47.0 and later), Opera (38.0 and later) or Google Chrome (51.0 and later) |
| Linux | Mozilla Firefox (47.0 and later), Opera (38.0 and later) |
| Mac | Mozilla Firefox (47.0 and later), Opera (38.0 and later) |
Top ↑
Popenda, M., Szachniuk, M., Antczak, M., Purzycka, K.J., Lukasiak, P., Bartol, N., Blazewicz, J., Adamiak, R.W. Automated 3D structure composition for large RNAs, Nucleic Acids Research, 2012, 40(14):e112 (doi:10.1093/nar/gks339).
Top ↑
RNAComposer maintains an official mirror site available at http://rnacomposer.cs.put.poznan.pl. Please, use RNAComposer mirror if the current site is overloaded, your job processing time is irrationally long, if you observe unstable system behaviour or network disruption problems.
Top ↑



